- Research Briefing
- Published:
(2025)Cite this article
Subjects
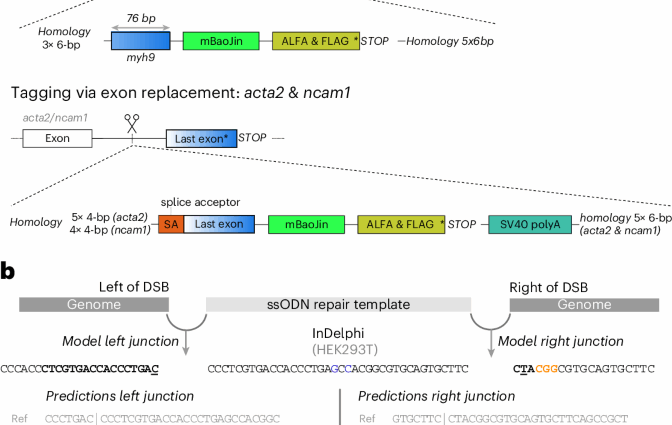
Precision CRISPR–Cas9-mediated genome engineering remains challenging, particularly gene integration and editing in non-dividing cells. We present Pythia, a deep learning solution that forecasts optimal repair templates and enables predictable and accurate genome editing in diverse cellular contexts, both in vivo and in vitro.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

References
-
Wang, J. Y. & Doudna, J. A. CRISPR technology: a decade of genome editing is only the beginning. Science 379, eadd8643 (2023). This review explores the rapid evolution of CRISPR genome editing over the past decade and outlines key advances, challenges and future directions across medicine, agriculture and basic research.
-
van Overbeek, M. et al. DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Mol. Cell 63, 633–646 (2016). This study demonstrates that DNA repair outcomes following Cas9 cleavage are non-random and largely determined by the target sequence.
-
Shen, M. W. et al. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature 563, 646–651 (2018). This study shows that template-free CRISPR–Cas9 editing can yield highly predictable repair outcomes at pathogenic loci, laying the groundwork for AI-driven models to anticipate editing results.
-
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019). This study introduces prime editing, which uses a Cas9–reverse transcriptase and pegRNA-encoded template to install precise edits without double-strand breaks or separate donor DNA.
-
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). This study introduces cytosine base editing using a Cas9 nickase–cytidine deaminase fusion that converts C>T (G>A) without double-stranded breaks or donor DNA.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This is a summary of: Naert, T. et al. Precise, predictable genome integrations by deep-learning-assisted design of microhomology-based templates. Nat. Biotechnol. https://doi.org/10.1038/s41587-025-02771-0 (2025).
Rights and permissions
About this article
Cite this article
Pythia provides deep learning-driven precision in CRISPR–Cas9 genome engineering.
Nat Biotechnol (2025). https://doi.org/10.1038/s41587-025-02818-2
-
Published:
-
DOI: https://doi.org/10.1038/s41587-025-02818-2